MethBase: a reference methylome database
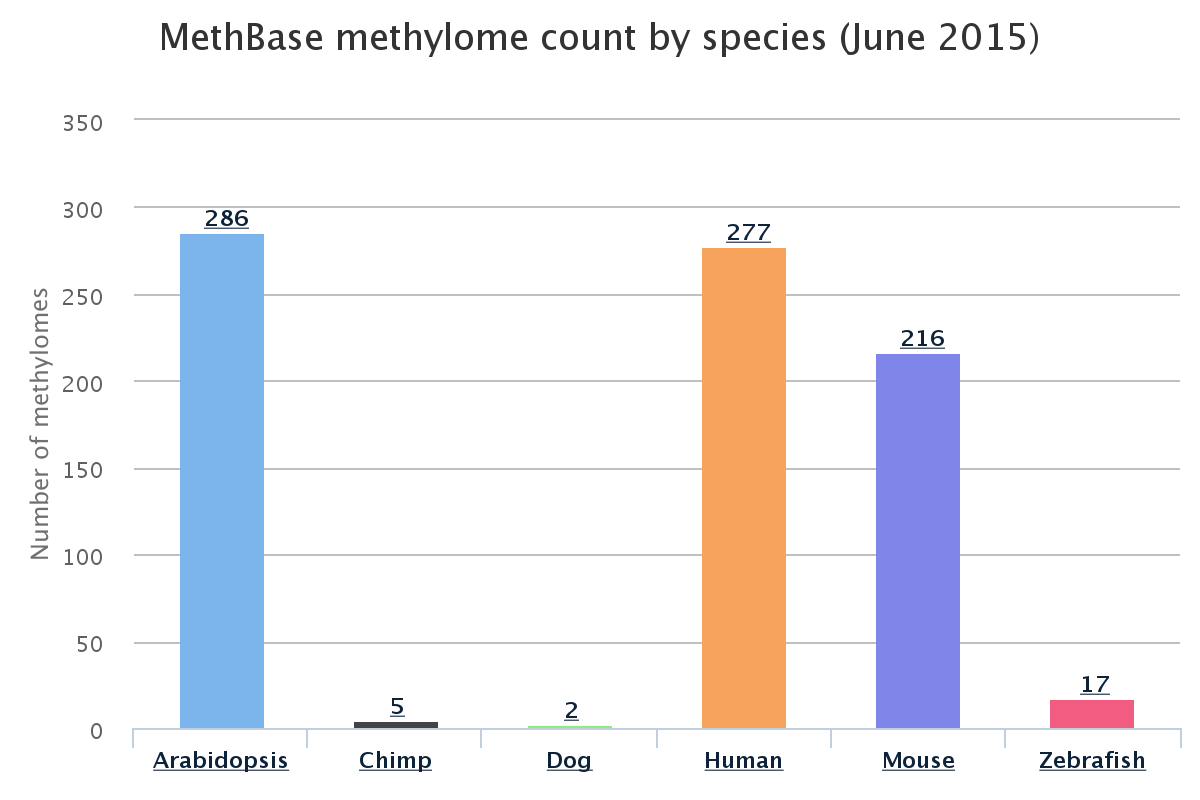
MethBase is a central reference methylome database created from public BS-seq datasets. It contains hundreds of methylomes from well studied organisms. For each methylome, Methbase provides methylation level at individual sites, regions of allele specific methylation, hypo- or hyper-methylated regions, partially methylated regions, and detailed meta data and summary statistics. These results are generated with the MethPipe software package, a standalone, comprehensive pipeline for analyzing bisulfite sequencing data, both WGBS and RRBS. You can obtain a pdf version of the MethBase Manual here.
Demo sessions
The following saved “sessions” pre-load a set of UCSC Genome Browser tracks that showcase some of the data from MethBase that can be viewed in the browser.
Human:
- Brain: This session shows tracks from brain tissue methylomes and neural progenitors.
- ESC/iPSC: This session shows a collection of tracks from ESCs and iPSCs. Also included: methylomes for cells that are origins of an iPSC or derived form an iPSC.
- Blood: This session shows a collection of tracks from multiple blood cells.
Mouse:
- Reprogramming: This session shows tracks from various stages of mouse male germ cell epigenomic reprogramming.
- mESCs: This session shows tracks from several mouse ESC methylomes produced by different groups in different projects.
- Blood: This session shows a collection of tracks from mouse blood cells.
To access MethBase
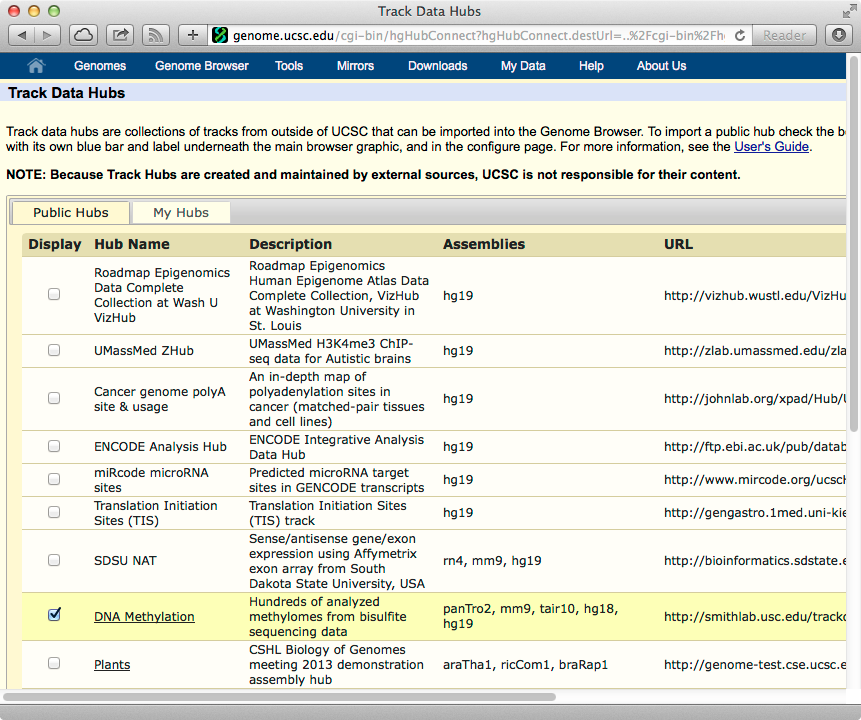
MethBase is publicly available to the scientific community as a track hub in the UCSC Genome Browser. If you are using the main site of UCSC Genome Browser, the MethBase track hub is built in by default, you can select the MethBase tracks from the Public Hubs section of the UCSC Genome Browser:
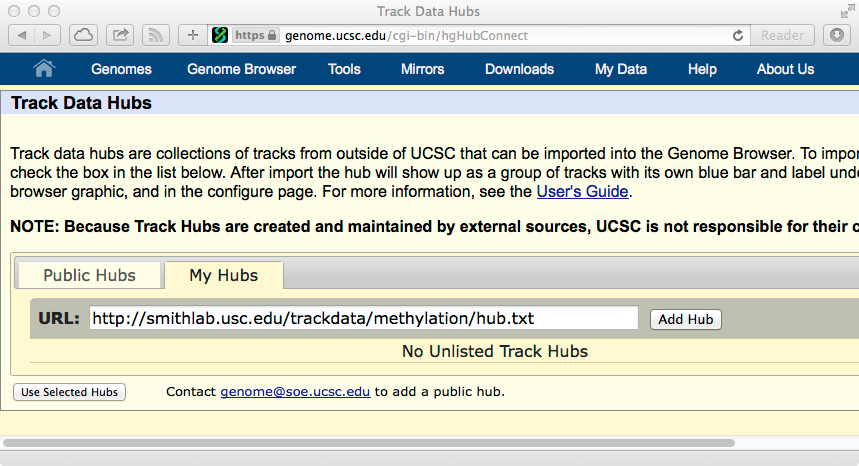
If you are using the a mirror site of UCSC Genome Browser, you can make the MethBase tracks available by loading the configuration file:
http://smithlab.usc.edu/trackdata/methylation/hub.txt
This is illustrated below. Please refer to Using UCSC Genome Browser Track Hubs for instructions on how to view unlisted hubs. The following shows how to add the MethBase track hub in mirror sites of UCSC Genome Browser:
Select tracks for viewing
After you load the MethBase track hub settings file and go to the Genome Browser page, it shows the methylation level tracks and the HMR tracks of a set of preselected methylomes. Below are examples of high-level methylation features available in MethBase through the UCSC genome browser track hub:
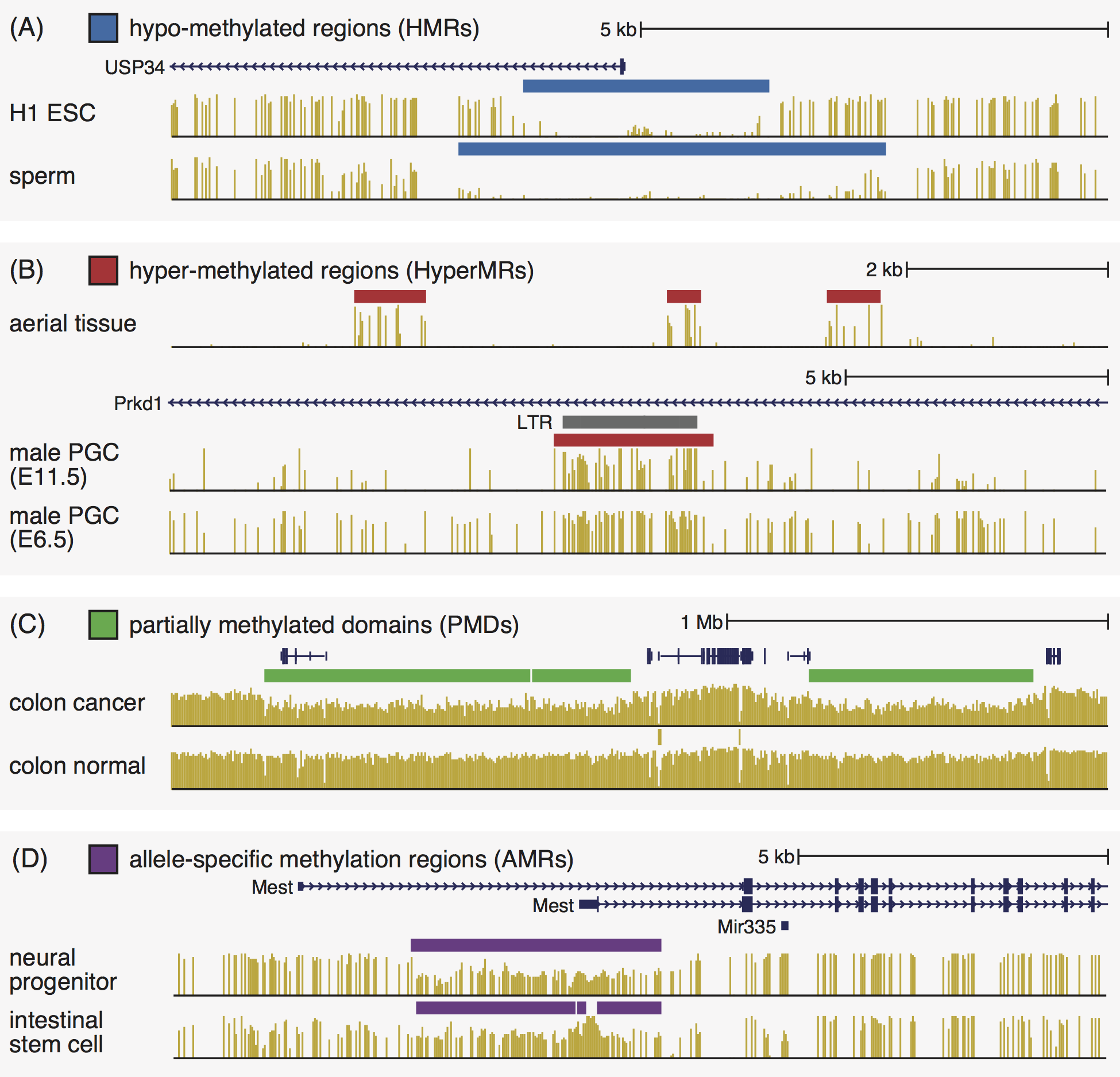
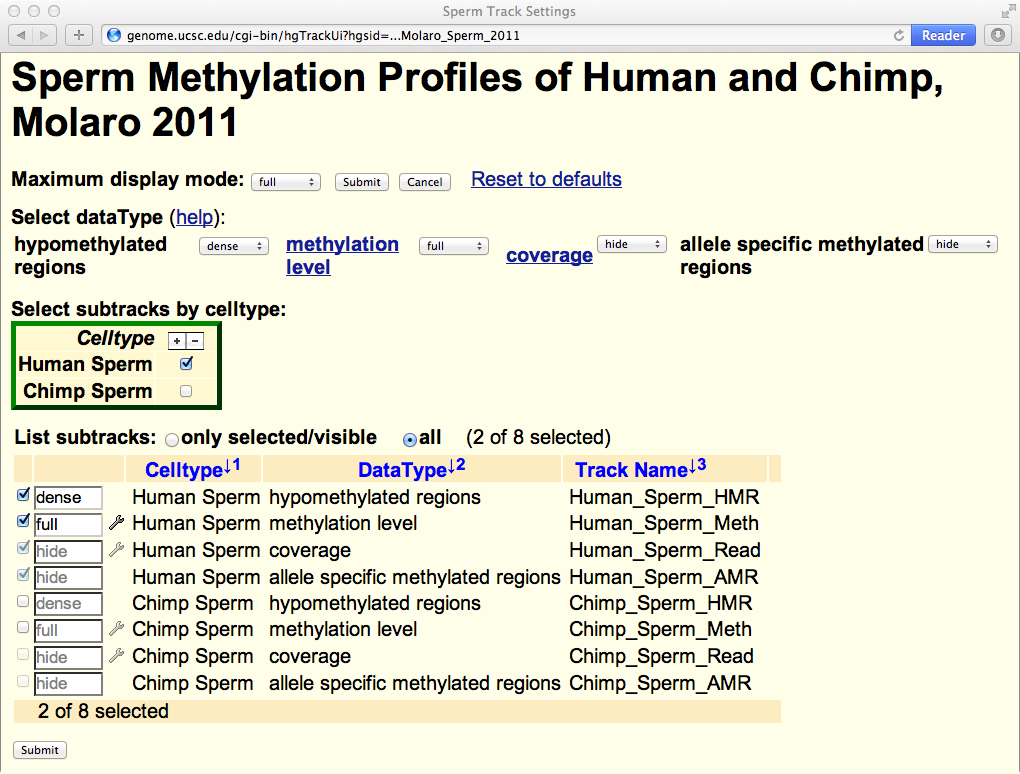
Besides the default methylation tracks and HMR tracks, MethBase provides a variety of other tracks, including coverage, hyper-methylated regions, partially methylated domains and allele-specific methylated AMRs (as shown above). To display those tracks, go to the track setting page in one of the three ways: 1) click the project name from the list of projects; 2) right click on of the viewable tracks, and select Configure track settings from the menu; or 3) go to the track description page and click the link Go to track controls. In the track setting page (shown below), you can select additional samples from that project and modify the display settings of more types of data.
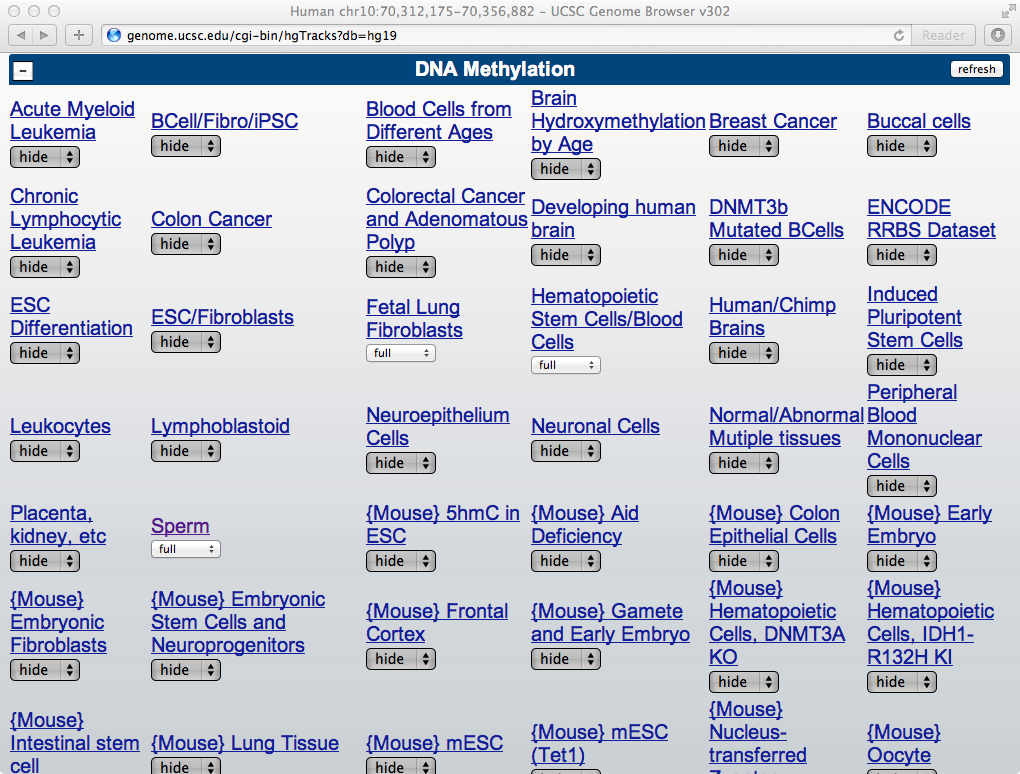
Below the Browser, you will a list of public methylomes in MethBase under the DNA Methylation section (shown below). From the list, you can modify the display settings of several samples from a project:
View meta data and summary statistics
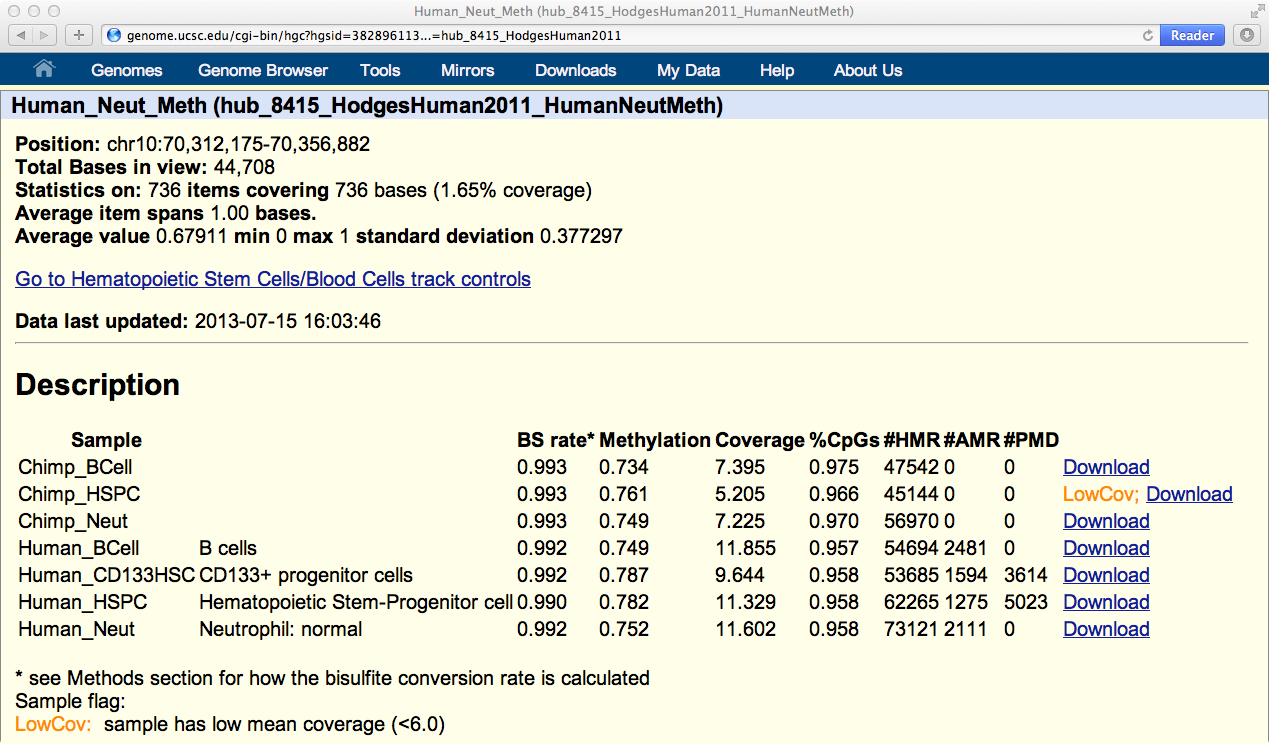
You can click on any track to go to the Description page for that track, which gives detailed meta and summary statistics about that methylome:
Downloading data for further analysis
If you use this resource, please cite us! Citations help enable us to keep generating this resource. The Smith Lab at USC has developed and is owner of the MethBase database, including all analyses and associated browser tracks (e.g. as displayed in the “DNA Methylation” trackhub on the UCSC Genome Browser). Any derivative work or use of the MethBase resource that appears in published literature must cite the most recent publication associated with MethBase (see “References” below). Users who wish to copy the contents of MethBase in bulk into a publicly available resource must additionally have explicit permission from the Smith Lab to do so. We hope the MethBase resource can help you!
Contact
Please send emails to the methpipe mailing list at methpipe@googlegroups.com if you have any questions, suggestions or comments. Visit MethBase Users Mailing List
References
Song Q, Decato B, Hong E, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD (2013) A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLOS ONE 8(12): e81148 [PDF] [Publisher Site]