MethPipe: a computational pipeline for analyzing bisulfite sequencing data
The MethPipe software package is a computational pipeline for analyzing bisulfite sequencing data (WGBS and RRBS).
Update July 2021: MethPipe now accepts SAM input after the read-mapping phase. Our old mr format is no longer supported. For short-read bisulfite mapping, we have a new tool called abismal. Download abismal-0.3.0 here.
Quick start
- Download source code: methpipe-5.0.0.tar.bz2 (release history)
- Read the manual: methpipe-manual.pdf
- Give it a try: Test data (17MB)
- View pre-analyzed methylomes: MethBase
- Check the latest development branch at GitHub Repo
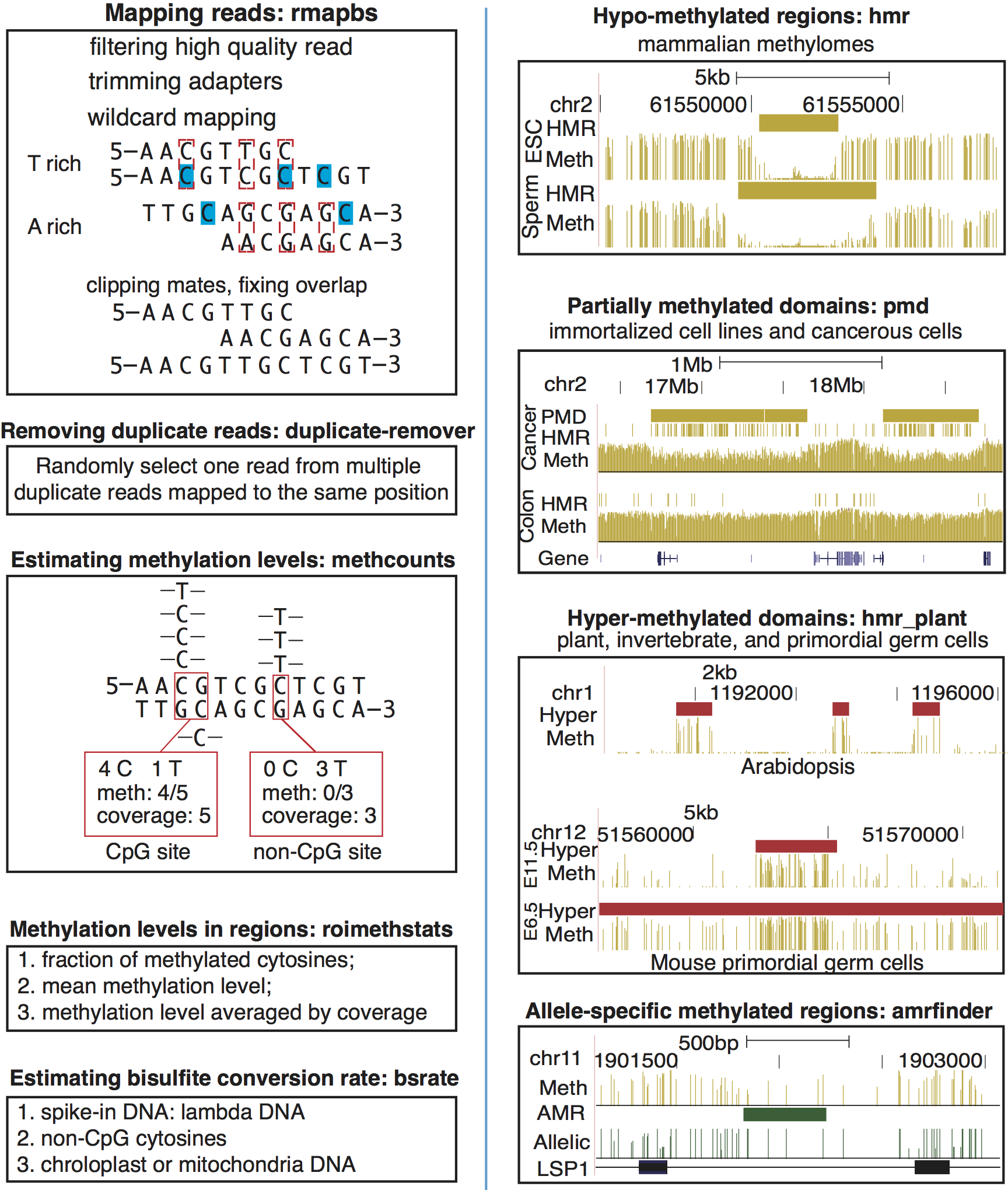
Tools
| Program | Description |
|---|---|
| abismal | map bisulfite treated short reads. See the Github repo for more details. |
| duplicate-remover | remove duplicate reads |
| methcounts | calculate methylation level and read coverage at individual sites |
| bsrate | estimate bisulfite conversion rate |
| hmr | identify hypo-methylated regions |
| hypermr | identify hyper-methylation in plants and organisms showing mosaic methylation |
| pmd | identify large partially methylated domains |
| amrfinder | identify allele-specific methylated regions |
| roimethstat | compute methylation level by genomic region of interest |
| mlml | consistently estimate 5-mC and 5-hmC levels. See MLML homepage. |
Workflow
Contact
Please send emails to the methpipe mailing list at
methpipe@googlegroups.com if you have any questions,
suggestions or comments.
To join the mailing list, send an email to methpipe+subscribe@googlegroups.com
View previous messages at MethPipe and MethBase Users’ Mailing List.
References
Song Q, Decato B, Hong E, Zhou M, Fang F, Qu J, Garvin T, Kessler M, Zhou J, Smith AD (2013) A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLOS ONE 8(12): e81148 [PDF] [Publisher Site]